Virtual Reality Inspection of Chromatin 3D and 2D Data
Description:
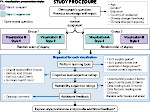
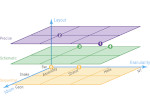
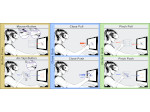
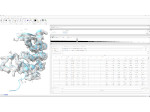
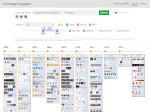
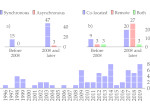
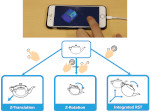
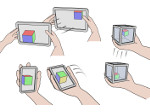
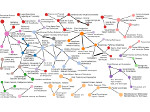
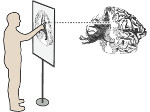
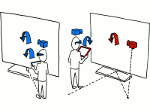
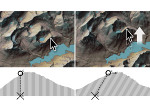
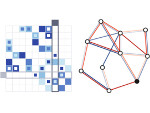
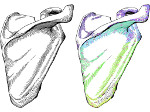
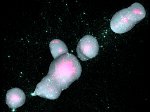
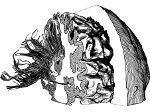
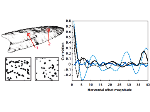
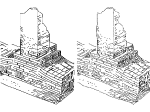
Understanding the packing of long DNA strands into chromatin is one of the ultimate challenges in genomic research. An intrinsic part of this complex problem is studying the chromatin’s spatial structure. Biologists reconstruct 3D models of chromatin from experimental data, yet the exploration and analysis of such 3D structures is limited in existing genomic data visualization tools. To improve this situation, we investigated the current options of immersive methods and designed a prototypical VR visualization tool for 3D chromatin models that leverages virtual reality to deal with the spatial data. We showcase the tool in three primary use cases. First, we provide an overall 3D shape overview of the chromatin to facilitate the identification of regions of interest and the selection for further investigation. Second, we include the option to export the selected regions and elements in the BED format, which can be loaded into common analytical tools. Third, we integrate epigenetic modification data along the sequence that influence gene expression, either as in-world 2D charts or overlaid on the 3D structure itself. We developed our application in collaboration with two domain experts and gathered insights from two informal studies with five other experts.
Paper download: 
 (31.0 MB)
(31.0 MB)
Video:
paper video:
Get the video:
Pictures:
(this image as well as others from the paper that are our own are available under a  CC-BY 4.0 license, see the license statement at the end of the paper)
CC-BY 4.0 license, see the license statement at the end of the paper)
Reference:
This work was done at and in collaboration with both the ViRVIG group at the Universitat Politècnica de Catalunya, Spain, and the Visitlab at Masaryk University, Czech Republic, as well as in collaboration with Harvard Medical School, USA.