Multiscale Visualization and Scale-Adaptive Modification of DNA Nanostructures
Description:
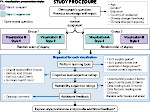
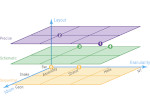
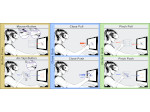
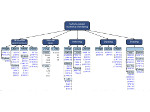
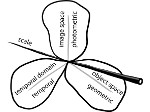
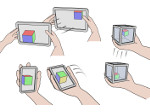
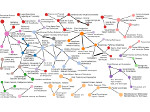
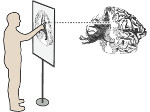
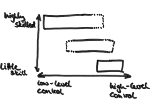
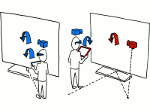
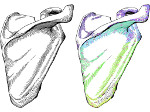
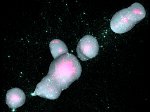
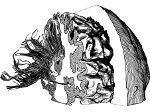
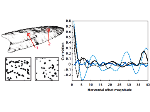
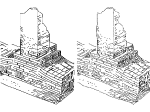
We present an approach to represent DNA nanostructures in varying forms of semantic abstraction, describe ways to smoothly transition between them, and thus create a continuous multiscale visualization and interaction space for applications in DNA nanotechnology. This new way of observing, interacting with, and creating DNA nanostructures enables domain experts to approach their work in any of the semantic abstraction levels, supporting both low-level manipulations and high-level visualization and modifications. Our approach allows them to deal with the increasingly complex DNA objects that they are designing, to improve their features, and to add novel functions in a way that no existing single-scale approach offers today. For this purpose we collaborated with DNA nanotechnology experts to design a set of ten semantic scales. These scales take the DNA's chemical and structural behavior into account and depict it from atoms to the targeted architecture with increasing levels of abstraction. To create coherence between the discrete scales, we seamlessly transition between them in a well-defined manner. We use special encodings to allow experts to estimate the nanoscale object's stability. We also add scale-adaptive interactions that facilitate the intuitive modification of complex structures at multiple scales. We demonstrate the applicability of our approach on an experimental use case. Moreover, feedback from our collaborating domain experts confirmed an increased time efficiency and certainty for analysis and modification tasks on complex DNA structures. Our method thus offers exciting new opportunities with promising applications in medicine and biotechnology.
Paper download:  (35.0 MB)
(35.0 MB)
Videos:
30 second preview:
paper presentation at IEEE  2017:
2017:
Get the videos:
- download the paper video (MPEG4, 170MB),
- watch the paper video on YouTube,
- watch the paper preview video on Vimeo,
- watch the paper presentation video on Vimeo.
Cross-Reference:
See our follow-up creation of an abstraction space for DNA nanostructures and our more general discussion of visual abstraction in (illustrative) visualization.
Main Reference:
Other Reference:
| Haichao Miao (2019) Geometric Abstraction for Effective Visualization and Modeling. PhD thesis, TU Wien, Austria, August 2019. | | ||
This work was done at and in collaboration with the Research Unit of Computer Graphics at TU Wien, Austria, as well as the Austrian Institute of Technology. Also see the AIT Website about the project as well as the page at TU Wien.
This work was supported through the Illustrare project, funded by ANR in France and FWF in Austria.