Moliverse: Contextually Embedding the Microcosm into the Universe
Description:
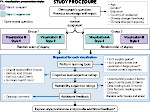
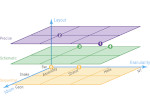
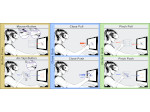
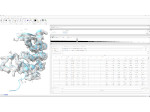
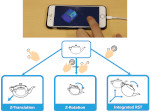
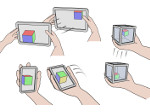
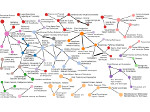
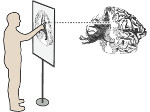
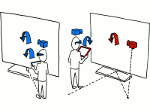
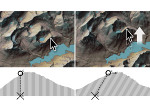
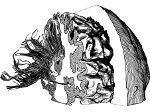
We present Moliverse, an integration of the molecular visualization framework VIAMD into the astronomical visualization software OpenSpace, allowing us to bridge the two extreme ends of the scale spectrum to show, for example, the gas composition in a planet's atmosphere or molecular structures in comet trails and can empower the creation of educational exhibitions. For that purpose we do not use a linear scale traversal but break the scale continuity and show molecular simulations as focus in the context of celestial bodies. We demonstrate the application of our concept in two storytelling scenarios and envision the application both for science presentations to lay audiences and for dedicated exploration, potentially also in a molecule-only environment.
Paper download: 
 (15.4 MB)
(15.4 MB)
Software:
The software is available at https://github.com/OpenSpace/OpenSpace/tree/feature/molecule_rendering (a branch in the OpenSpace repository that has ViaMD as a submodule).
Video:
paper video:
Get the video:
Pictures:
(these images as well as others in the paper are available under a  CC-BY 4.0 license, see the license statement at the end of the paper)
CC-BY 4.0 license, see the license statement at the end of the paper)
Reference:
This work was done at and in collaboration with the Visualization Centre of Linköping University, Sweden.