DiffFit: Visually-Guided Differentiable Fitting of Molecule Structures to a Cryo-EM Map
Description:
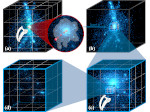
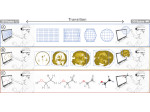
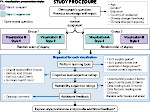
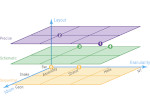
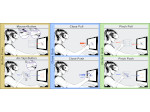
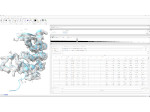
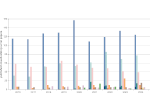
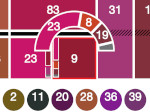
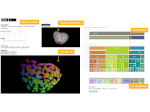
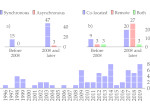
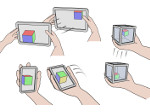
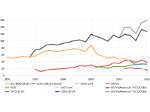
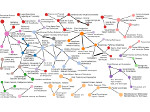
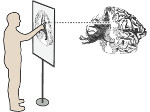
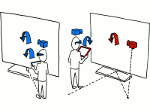
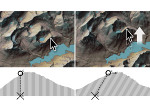
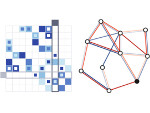
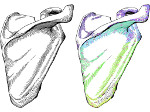
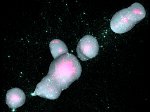
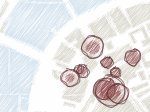
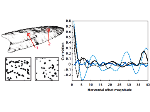
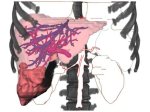
We introduce DiffFit, a differentiable algorithm for fitting protein atomistic structures into an experimental reconstructed Cryo-Electron Microscopy (cryo-EM) volume map. In structural biology, this process is necessary to semi-automatically composite large mesoscale models of complex protein assemblies and complete cellular structures that are based on measured cryo-EM data. The current approaches require manual fitting in three dimensions to start, resulting in approximately aligned structures followed by an automated fine-tuning of the alignment. The DiffFit approach enables domain scientists to fit new structures automatically and visualize the results for inspection and interactive revision. The fitting begins with differentiable three-dimensional (3D) rigid transformations of the protein atom coordinates followed by sampling the density values at the atom coordinates from the target cryo-EM volume. To ensure a meaningful correlation between the sampled densities and the protein structure, we proposed a novel loss function based on a multi-resolution volume-array approach and the exploitation of the negative space. This loss function serves as a critical metric for assessing the fitting quality, ensuring the fitting accuracy and an improved visualization of the results. We assessed the placement quality of DiffFit with several large, realistic datasets and found it to be superior to that of previous methods. We further evaluated our method in two use cases: automating the integration of known composite structures into larger protein complexes and facilitating the fitting of predicted protein domains into volume densities to aid researchers in identifying unknown proteins. We implemented our algorithm as an open-source plugin in ChimeraX, a leading visualization software in the field.
Errata:
Please note that an errata for this paper has been published in the journal. The PDFs available here are the updated version of the paper with slightly changed numbers and images, all integrated into the actual paper. For the specific changes see the errata PDF linked below.
Paper download: 
 (6.8 MB)
(6.8 MB)
Additional material:
We make several items of additional material available in the following OSF repository: osf.io/5tx4q.
Software:
The tool source code is also available at github.com/nanovis/DiffFitViewer. In addition, the tool has been integrated into the ChimeraX software infrastructure.
Videos:
paper video:
30s preview video for IEEE  :
:
pre-recorded presentation video for IEEE  :
:
presentation at IEEE  :
:
Get the videos:
- watch the paper video on YouTube
- download the paper video (MPEG4, 77.6MB)
- watch the 30s preview video on YouTube
- watch the pre-recorded presentation video on YouTube
- watch the actual presentation video on YouTube
Pictures:
(these images as well as others from the paper that are our own are available under a  CC-BY 4.0 license, see the license statement at the end of the paper)
CC-BY 4.0 license, see the license statement at the end of the paper)
Poster presented at the ICML 2024 Differentiable Almost Everything workshop:
Main Reference:
Other References:
| Deng Luo (2025) Visual Analytics for Macromolecular Science. PhD thesis, King Abdullah University of Science and Technology (KAUST), Saudi-Arabia, August 2025. | | ||
This work was done at and in collaboration with the Visual Computing Center of KAUST, Kingdom of Saudi-Arabia.