SynopFrame: Multiscale Time-Dependent Visual Abstraction Framework for Analyzing DNA Nanotechnology Simulations
Description:
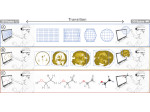
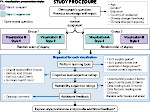
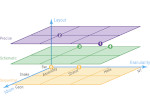
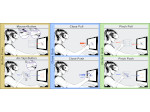
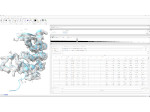
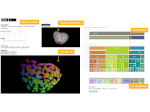
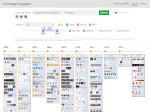
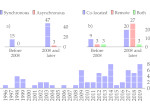
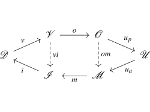
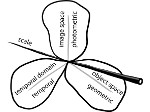
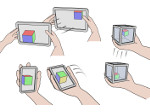
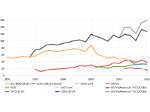
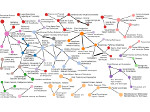
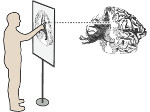
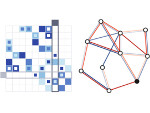
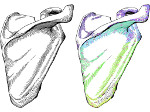
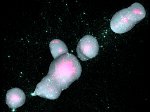
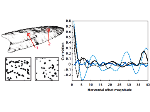
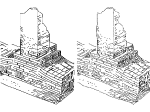
We present an open-source framework, SynopFrame, that allows DNA nanotechnology (DNA-nano) experts to analyze and understand molecular dynamics simulation trajectories of their designs. We use a multiscale multi-dimensional abstraction space, connect the representations to a projected conformational space plot of the structure’s temporal sequence, and thus enable experts to analyze the dynamics of their structural designs and, specifically, failure cases of the assembly. In addition, our time-dependent abstraction representation allows the biologists, for the first time in a smooth and structurally clear way, to identify and observe temporal transitions of a DNA-nano design from one configuration to another, and to highlight important periods of the simulation for further analysis. We realize SynopFrame as a dashboard of the different synchronized 3D spatial and 2D schematic visual representations, with a color overlay to show essential properties such as the status of hydrogen bonds. The linking of the spatial, schematic, and abstract views ensures that users can effectively analyze the high-frequency motion. We also categorize the status of the hydrogen bonds into a new format to allow us to color-encode it and overlay it on the representations. To demonstrate the utility of SynopFrame, we describe example usage scenarios and report user feedback.
Paper download: coming soon
Additional material:
We make several items of additional material available in the following OSF repository: osf.io/qxrzw.
Software:
The tool source code is also available at https://github.com/nanovis/SynopFrame.
Video:
paper video:
Get the video:
Pictures:
(this image as well as others from the paper that are our own are available under a  CC-BY 4.0 license, see the license statement at the end of the paper)
CC-BY 4.0 license, see the license statement at the end of the paper)
Main Reference:
Other reference:
| Deng Luo (2025) Visual Analytics for Macromolecular Science. PhD thesis, King Abdullah University of Science and Technology (KAUST), Saudi-Arabia, August 2025. | | ||
This work was done at and in collaboration with the Visual Computing Center of KAUST, Kingdom of Saudi-Arabia.