LineageD: An Interactive Visual System for Plant Cell Lineage Assignments based on Correctable Machine Learning
Description:
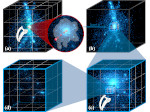
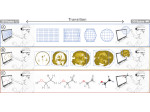
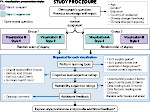
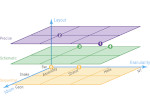
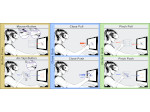
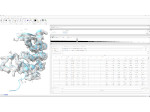
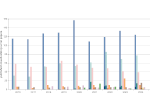
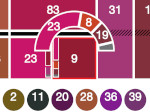
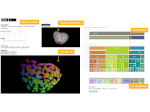
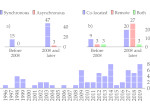
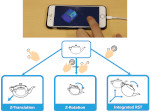
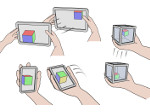
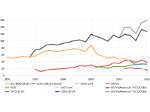
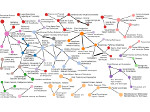
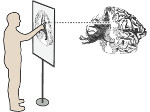
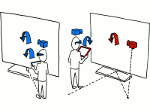
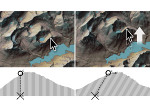
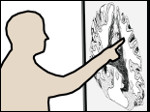
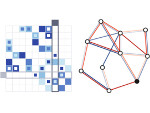
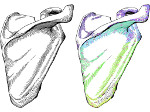
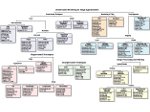
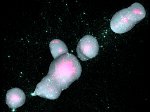
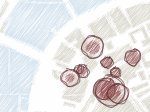
We describe LineageD—a hybrid web-based system to predict, visualize, and interactively adjust plant embryo cell lineages. Currently, plant biologists explore the development of an embryo and its hierarchical cell lineage manually, based on a 3D dataset that represents the embryo status at one point in time. This human decision-making process, however, is time-consuming, tedious, and error-prone due to the lack of integrated graphical support for specifying the cell lineage. To fill this gap, we developed a new system to support the biologists in their tasks using an interactive combination of 3D visualization, abstract data visualization, and correctable machine learning to modify the proposed cell lineage. We use existing manually established cell lineages to obtain a neural network model. We then allow biologists to use this model to repeatedly predict assignments of a single cell division stage. After each hierarchy level prediction, we allow them to interactively adjust the machine learning based assignment, which we then integrate into the pool of verified assignments for further predictions. In addition to building the hierarchy this way in a bottom-up fashion, we also offer users to divide the whole embryo and create the hierarchy tree in a top-down fashion for a few steps, improving the ML-based assignments by reducing the potential for wrong predictions. We visualize the continuously updated embryo and its hierarchical development using both 3D spatial and abstract tree representations, together with information about the model's confidence and spatial properties. We conducted case study validations with five expert biologists to explore the utility of our approach and to assess the potential for LineageD to be used in their daily workflow. We found that the visualizations of both 3D representations and abstract representations help with decision making and the hierarchy tree top-down building approach can reduce assignments errors in real practice.
Paper download:  (12.0 MB)
(12.0 MB)
Software:
The software to set up a local copy of the server tool is available at https://gitlab.inria.fr/jhong/lineaged.
Demo:
We also have an online demo of LineageD. For getting the password, please contact us via e-mail.
Study materials and data:
Our study materials and data can be found in the following OSF repository: osf.io/rhyg4.
Videos:
Get the videos:
Pictures:
(these images as well as others in the paper are available under a  CC-BY 4.0 license, see the license statement at the end of the paper)
CC-BY 4.0 license, see the license statement at the end of the paper)
Cross-Reference:
We continued this project, which led to our work on LineageD+. Please check this out as well.
Main Reference:
Other Reference:
This work was done at the AVIZ project group of Inria, France, in collaboration with the MaiAGE team of InraE Jouy-en-Josas. There is also a separate project webpage on the Aviz server.
This work was supported through the Naviscope project, funded by Inria, France.