Visualizing and Comparing Machine Learning Predictions to Improve Human-AI Teaming on the Example of Cell Lineage
Description:
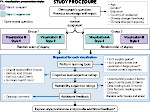
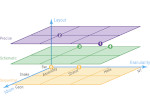
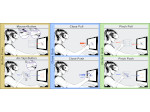
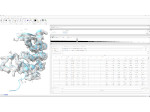
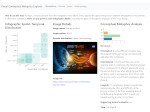
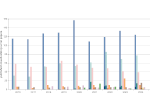
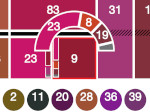
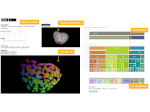
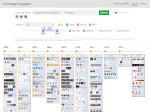
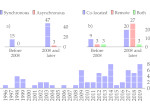
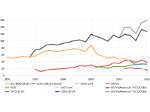
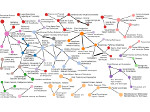
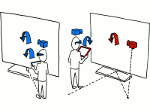
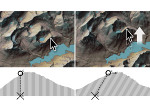
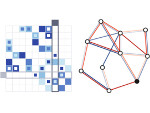
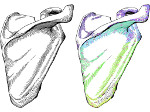
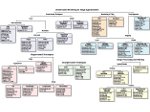
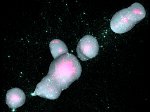
We visualize the predictions of multiple machine learning models to help biologists as they interactively make decisions about cell lineage—the development of a (plant) embryo from a single ovum cell. Based on a confocal microscopy dataset, traditionally biologists manually constructed the cell lineage, starting from this observation and reasoning backward in time to establish their inheritance. To speed up this tedious process, we make use of machine learning (ML) models trained on a database of manually established cell lineages to assist the biologist in cell assignment. Most biologists, however, are not familiar with ML, nor is it clear to them which model best predicts the embryo’s development. We thus have developed a visualization system that is designed to support biologists in exploring and comparing ML models, checking the model predictions, detecting possible ML model mistakes, and deciding on the most likely embryo development. To evaluate our proposed system, we deployed our interface with six biologists in an observational study. Our results show that the visual representations of machine learning are easily understandable, and our tool, LineageD+, could potentially increase biologists’ working efficiency and enhance the understanding of embryos.
Paper download:  (13.0 MB)
(13.0 MB)
Study materials and data:
Our study materials and data can be found in the following OSF repository: osf.io/dcek9. Our study pre-registration is at osf.io/2f6uc.
Software:
The software is available at github.com/JiayiHong/LineageD_Plus.
Demo:
We also have an online demo of LineageD+. For getting the password, please contact us via e-mail.
Videos:
paper video:
30s preview video for IEEE  :
:
pre-recorded presentation video for IEEE  :
:
presentation at IEEE  :
:
Get the videos:
- watch the paper video on YouTube
- download the paper video (MPEG4, 146MB)
- watch the 30s preview video on YouTube
- watch the pre-recorded presentation video on YouTube
- watch the actual presentation video on YouTube
Pictures:
(these images as well as others from the paper are available under a  CC-BY 4.0 license, see the license statement at the end of the paper)
CC-BY 4.0 license, see the license statement at the end of the paper)
Cross-Reference:
This project is based on our earlier work on LineageD. Please check this out as well.
Main Reference:
Other Reference:
This work was done at the AVIZ project group of Inria, France.
This work was supported through the Naviscope project, funded by Inria, France.